Nature Methods on Twitter: "Avant-garde: a tool for refining DIA-mass spectrometry data. @JaffeLab https://t.co/zIWJ9PUYiz… "

Simultaneous Improvement in the Precision, Accuracy, and Robustness of Label-free Proteome Quantification by Optimizing Data Manipulation Chains | Molecular & Cellular Proteomics

Hybrid Spectral Library Combining DIA-MS Data and a Targeted Virtual Library Substantially Deepens the Proteome Coverage - ScienceDirect

Group-DIA: analyzing multiple data-independent acquisition mass spectrometry data files | Nature Methods

Machine Learning in Mass Spectrometric Analysis of DIA Data - Xu - - PROTEOMICS - Wiley Online Library
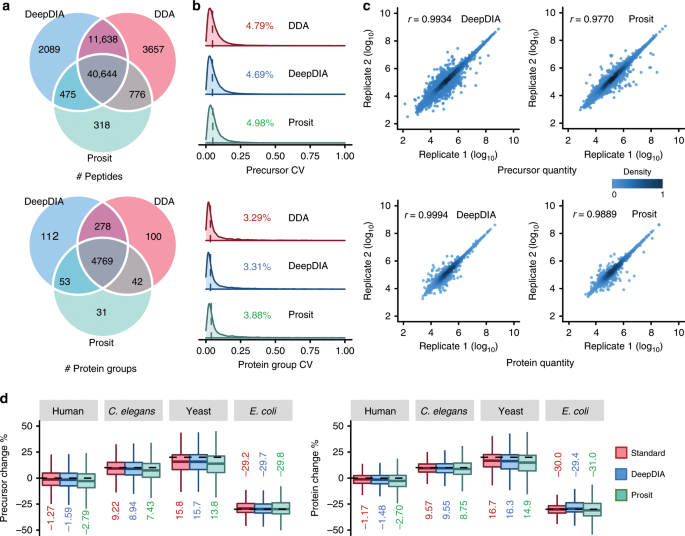
In silico spectral libraries by deep learning facilitate data-independent acquisition proteomics | Nature Communications
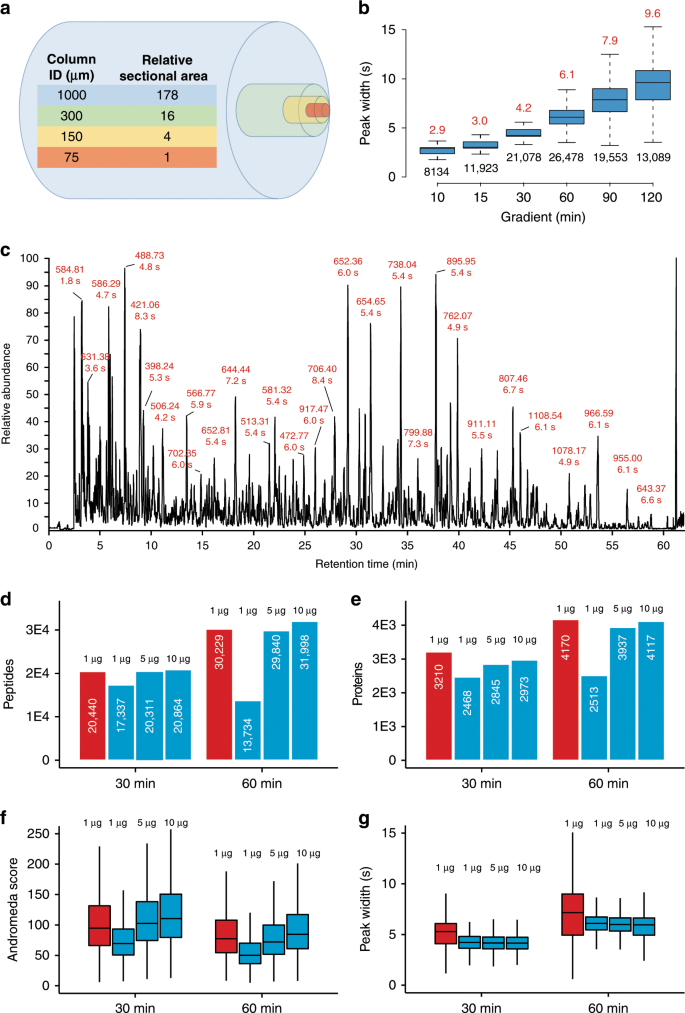
Robust, reproducible and quantitative analysis of thousands of proteomes by micro-flow LC–MS/MS | Nature Communications
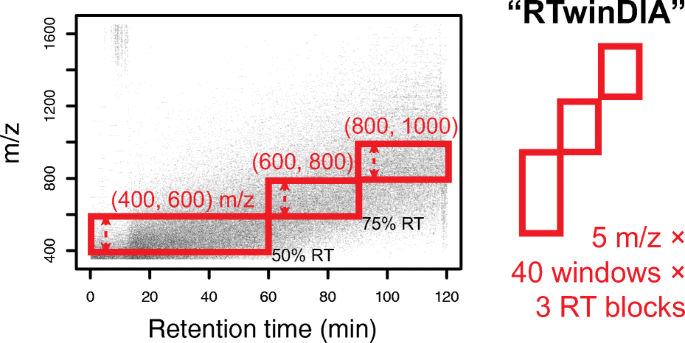
Assessing the Relationship Between Mass Window Width and Retention Time Scheduling on Protein Coverage for Data-Independent Acquisition | SpringerLink
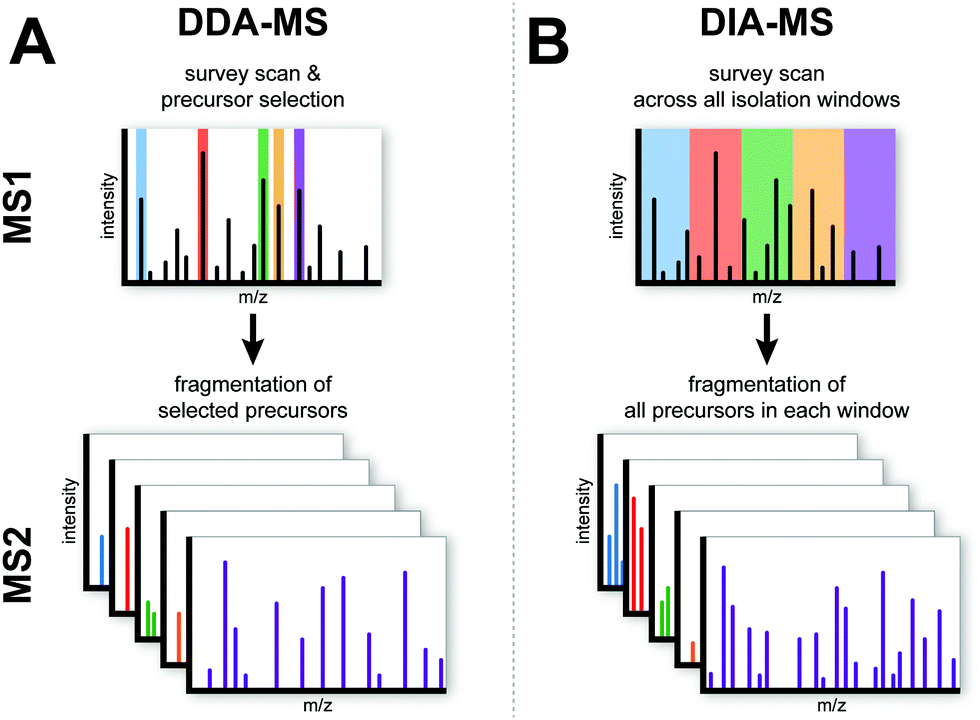
Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology - Molecular Omics (RSC Publishing) DOI:10.1039/D0MO00072H
Optimization of Experimental Parameters in Data-Independent Mass Spectrometry Significantly Increases Depth and Reproducibility

Deep learning enables de novo peptide sequencing from data-independent-acquisition mass spectrometry | Nature Methods
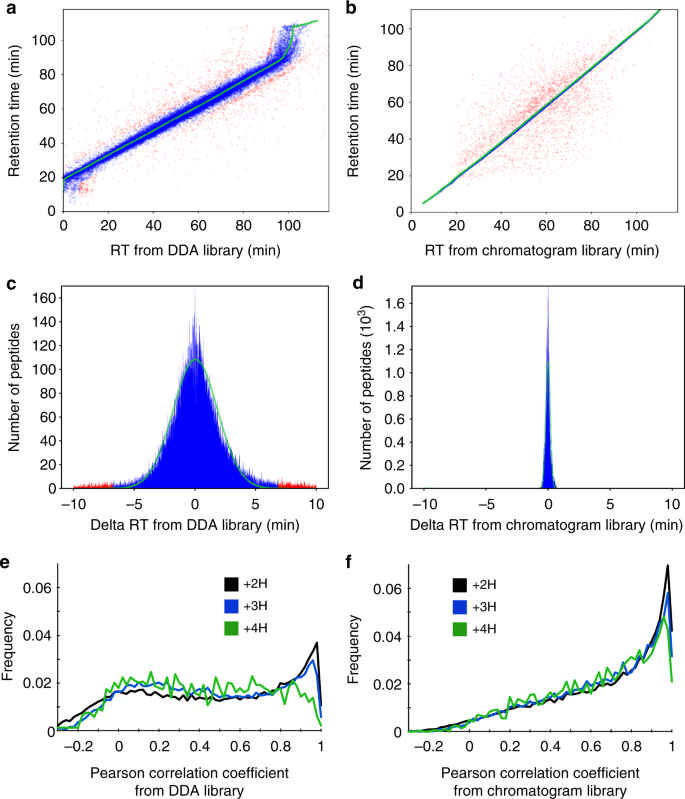
Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry | Nature Communications
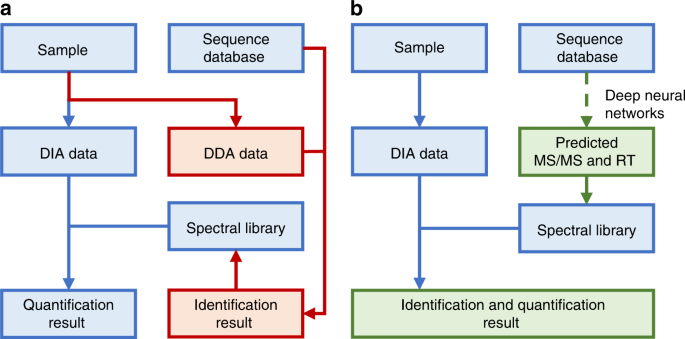
In silico spectral libraries by deep learning facilitate data-independent acquisition proteomics | Nature Communications

PulseDIA: in-depth data independent acquisition mass spectrometry using enhanced gas phase fractionation | bioRxiv